Public Data Repositories
What are data repositories?
Section titled “What are data repositories?”Public data repositories are one option to publish your research data. They usually focus on the data – as opposed to other research outputs such as manuscripts. Data repositories assign persistent identifiers (e.g. a DOI) to your dataset and by that comply with requirements of most publication journals.
We differentiate between domain-specific and general-purpose repositories.
Domain-specific data repositories
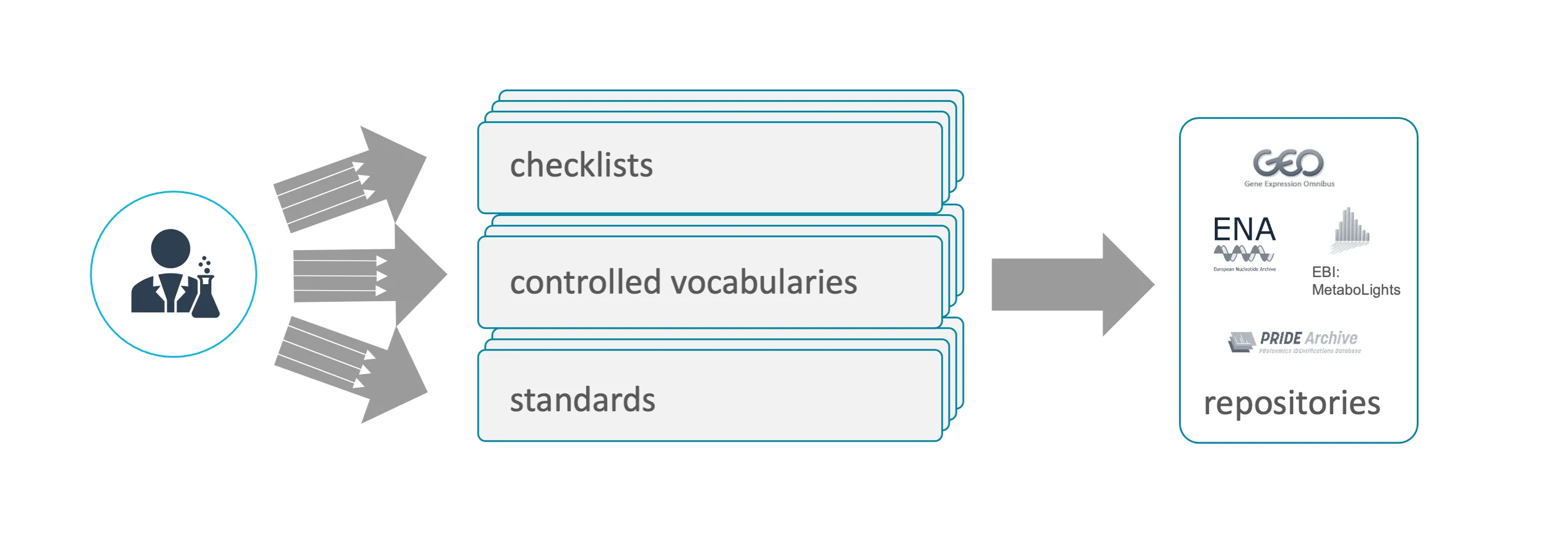
Section titled “Domain-specific data repositories”Domain-specific data repositories are well-established in a domain or community specialized on a certain data type. They frequently co-develop or foster compliance with metadata standards (see metadata) and oftentimes curate data. Data deposition at these repositories is recommended.
The following table lists examples of relevant endpoint repositories for data produced by DataPLANT participants. To simplify data submission, DataPLANT provides metadata templates for many of these repositories in Swate. Check the links below for additional repositories.
| Repository | Description | Biological data domain | DataPLANT Templates available |
|---|---|---|---|
| EBI-ENA | European Nucleotide Archive | genome / transcriptome sequences | |
| EBI-ArrayExpress | Archive of Functional Genomics Data | transcriptome | |
| EBI-MetaboLights | Database of Metabolomics | metabolome | |
| EBI-PRIDE | PRoteomics IDEntifications Database | proteome | |
| EBI-BioImage Archive | Stores and distributes biological images | imaging, microscopy | |
| e!DAL-PGP | Plant Genomics & Phenomics Research Data Repository | phenome | |
| NCBI-GEO | Gene Expression Omnibus | transcriptome | |
| NCBI-GenBank | Genetic Sequence Database | genome | |
| NCBI-SRA | Sequence Read Archive | genome / transcriptome sequences |
General-purpose repositories
Section titled “General-purpose repositories”In cases where no suitable domain-specific repository exists, general-purpose repositories are an option to publicly deposit research data and receive a PID. A benefit of general-purpose repositories is that they allow deposition of virtually any data type. Also research data packages with mixes of data types and computational workflows can be deposited, which aligns well with typical plant science investigations. However, since these repositories can only foster compliance with metadata standards at a very generic level (e.g. bibliographic or technical, see metadata), they limit the capacity for FAIR reuse of data.
Examples for general-purpose repositories include
- Zenodo https://zenodo.org,
- DRYAD https://datadryad.org/, and
- FigShare https://figshare.com.
Finding a suitable repository
Section titled “Finding a suitable repository”The following resources provide good starting points to seek a suitable repository for your research data.
- DataPLANT’s Metadata recommendation quiz
- FAIRsharing: https://fairsharing.org
- re3data (Registry of Research Data Repositories): https://www.re3data.org
- Overview of EMBL-EBI repositories: https://www.ebi.ac.uk/services/all
- Overview of NCBI repositories: https://www.ncbi.nlm.nih.gov/guide/sitemap/
Submitting data to a public data repository
Section titled “Submitting data to a public data repository”Depositing research data at a public data repository can be tedious. Especially the domain-specific repositories require compliance with specific data submission routines (a) in terms of format and content and (b) for both “raw data” and “metadata”. Only data types relevant for the respective domain are accepted and need to be provided in proper data formats. In order to guarantee that the information required to properly describe the data is present, they require adherence to domain-specific metadatastandards, represented in the proper format and oftentimes require the use of controlled vocabularies and ontologies. And finally the mere technicalities of how to collect and submit the (meta)data varies greatly between repositories, ranging from the use of pure upload via file transfers (e.g. FTP), APIs, online web forms or specialized software requiring local installation. The large repository providers invest a lot to harmonize their formats and submission routines. Still, there is a long way to go and we are currently far away from the unified way where “If you know one, you know them all.”