Annotation Tables
The Tables shown in xlsx files in ISA files are modelled in ARCtrl as ArcTable object.
These objects are assigned as ArcTables interface in both ArcAssay and ArcStudy.
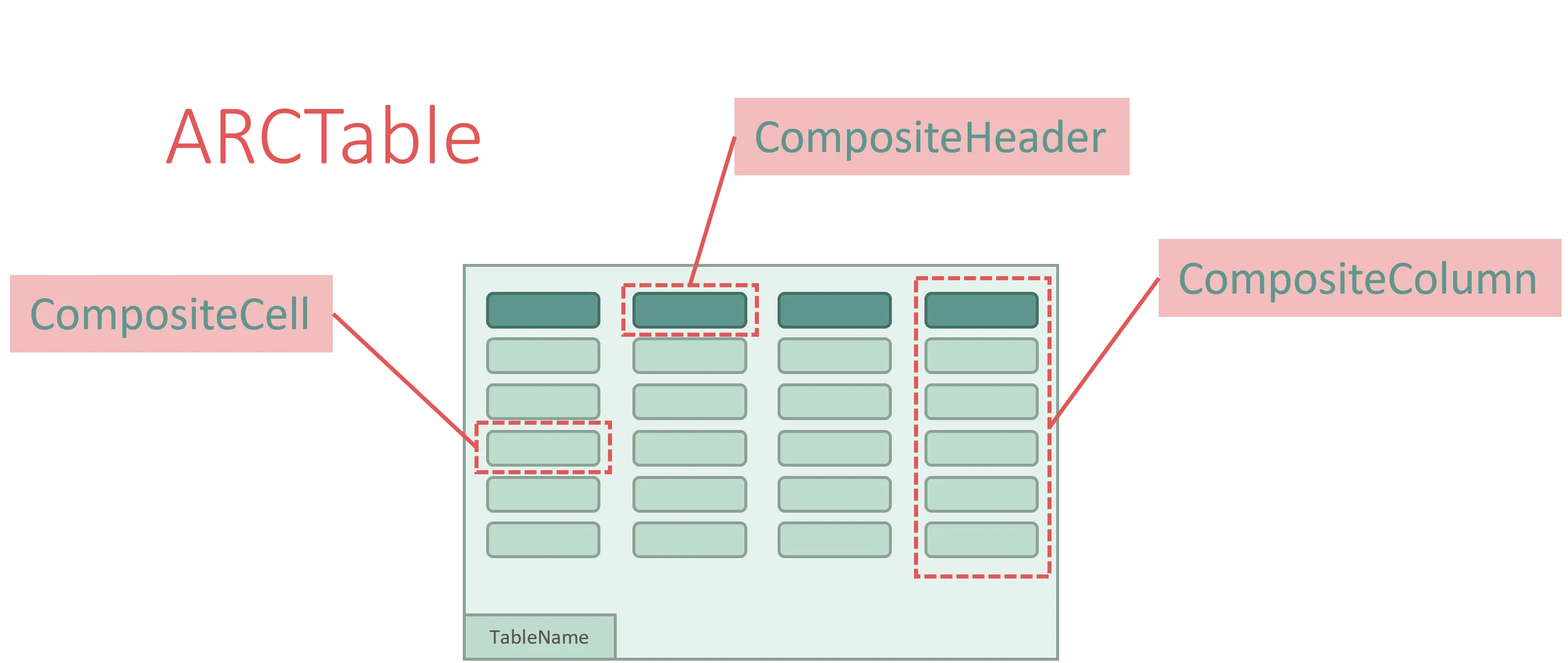
ArcTables are column-major objects, with the columns being typed as CompositeColumn.
Each CompositeColumn consists of a CompositeHeader and a collection of CompositeCells, which make up the values of the table.
Building an ArcTable
Section titled “Building an ArcTable”open ARCtrl
let growth = ArcTable.init("Growth")
// create ontology annotation for "species"let oa_species = OntologyAnnotation( "species", "NCIT", "NCIT:C45293" )// create ontology annotation for "chlamy"let oa_chlamy = OntologyAnnotation( "Chlamydomonas reinhardtii", "NCBITaxon", "NCBITaxon_3055" )
let oa_time = OntologyAnnotation( "time", "EFO", "EFO:0000721" )
let oa_day = OntologyAnnotation( "day", "UO", "UO:0000033" )
// This will create an input column with one row cell.// In xlsx this will be exactly 1 column.growth.AddColumn( CompositeHeader.Input IOType.Source, [|CompositeCell.createFreeText "Input1"|])
// this will create an Characteristic [species] column with one row cell.// in xlsx this will be exactly 3 columns.growth.AddColumn( CompositeHeader.Characteristic oa_species, [|CompositeCell.createTerm oa_chlamy|])
// this will create an Parameter [time] column with one row cell.// in xlsx this will be exactly 4 columns.growth.AddColumn( CompositeHeader.Parameter oa_species, [|CompositeCell.createUnitized("5", oa_day)|])
growth.AddColumn( CompositeHeader.Output IOType.Sample, [|CompositeCell.createFreeText "Output1"|])import {ArcTable, OntologyAnnotation, CompositeHeader, CompositeCell, IOType} from "@nfdi4plants/arctrl";
let growth = ArcTable.init("Growth")
let oa_species = new OntologyAnnotation("species", "NCIT", "NCIT:C45293")
let oa_chlamy = new OntologyAnnotation("Chlamydomonas reinhardtii", "NCBITaxon", "NCBITaxon_3055")
let oa_time = new OntologyAnnotation("time", "EFO", "EFO:0000721")
let oa_day = new OntologyAnnotation("day", "UO", "UO:0000033")
// This will create an input column with one row cell.// In xlsx this will be exactly 1 column.growth.AddColumn(CompositeHeader.input(IOType.source()), [CompositeCell.createFreeText("Input1")])
// This will create an Characteristic [species] column with one row cell.// In xlsx this will be exactly 3 columns.growth.AddColumn(CompositeHeader.characteristic(oa_species), [CompositeCell.createTerm(oa_chlamy)])
// This will create an Parameter [time] column with one row cell.// In xlsx this will be exactly 4 columns.growth.AddColumn(CompositeHeader.parameter(oa_time), [CompositeCell.createUnitized("5", oa_day)])
// This will create an Output column with one row cell.// In xlsx this will be exactly 1 columns.growth.AddColumn(CompositeHeader.output(IOType.sample()), [CompositeCell.createFreeText("Output1")])from arctrl.arctrl import ArcTable, OntologyAnnotation, CompositeHeader, CompositeCell, IOType
growth = ArcTable.init("Growth")
oa_species = OntologyAnnotation("species", "NCIT", "NCIT:C45293")
oa_chlamy = OntologyAnnotation("Chlamydomonas reinhardtii", "NCBITaxon", "NCBITaxon_3055")
oa_time = OntologyAnnotation("time", "EFO", "EFO:0000721")
oa_day = OntologyAnnotation("day", "UO", "UO:0000033")
# This will create an input column with one row cell.# In xlsx this will be exactly 1 column.growth.AddColumn(CompositeHeader.input(IOType.source()), [CompositeCell.create_free_text("Input1")])
# This will create an Characteristic [species] column with one row cell.# In xlsx this will be exactly 3 columns.growth.AddColumn(CompositeHeader.characteristic(oa_species), [CompositeCell.create_term(oa_chlamy)])
# This will create an Parameter [time] column with one row cell.# In xlsx this will be exactly 4 columns.growth.AddColumn(CompositeHeader.parameter(oa_time), [CompositeCell.create_unitized("5", oa_day)])
# This will create an Output column with one row cell.# In xlsx this will be exactly 1 columns.growth.AddColumn(CompositeHeader.output(IOType.sample()), [CompositeCell.create_free_text("Output1")])