sugar_result.csv
| sug_s1_r1 | sug_s1_r2 |
|---|---|
| 15.5 | 14.8 |
| 15.2 | 15.1 |
| 15.9 | 14.7 |
The Datamap files used to annotate data files are modeled in ARCtrl as Datamap objects.
They can be found in Study, Assay, Workflow and Run folders and therefore in the ArcStudy, ArcAssay, ArcWorkflow and ArcRun classes.
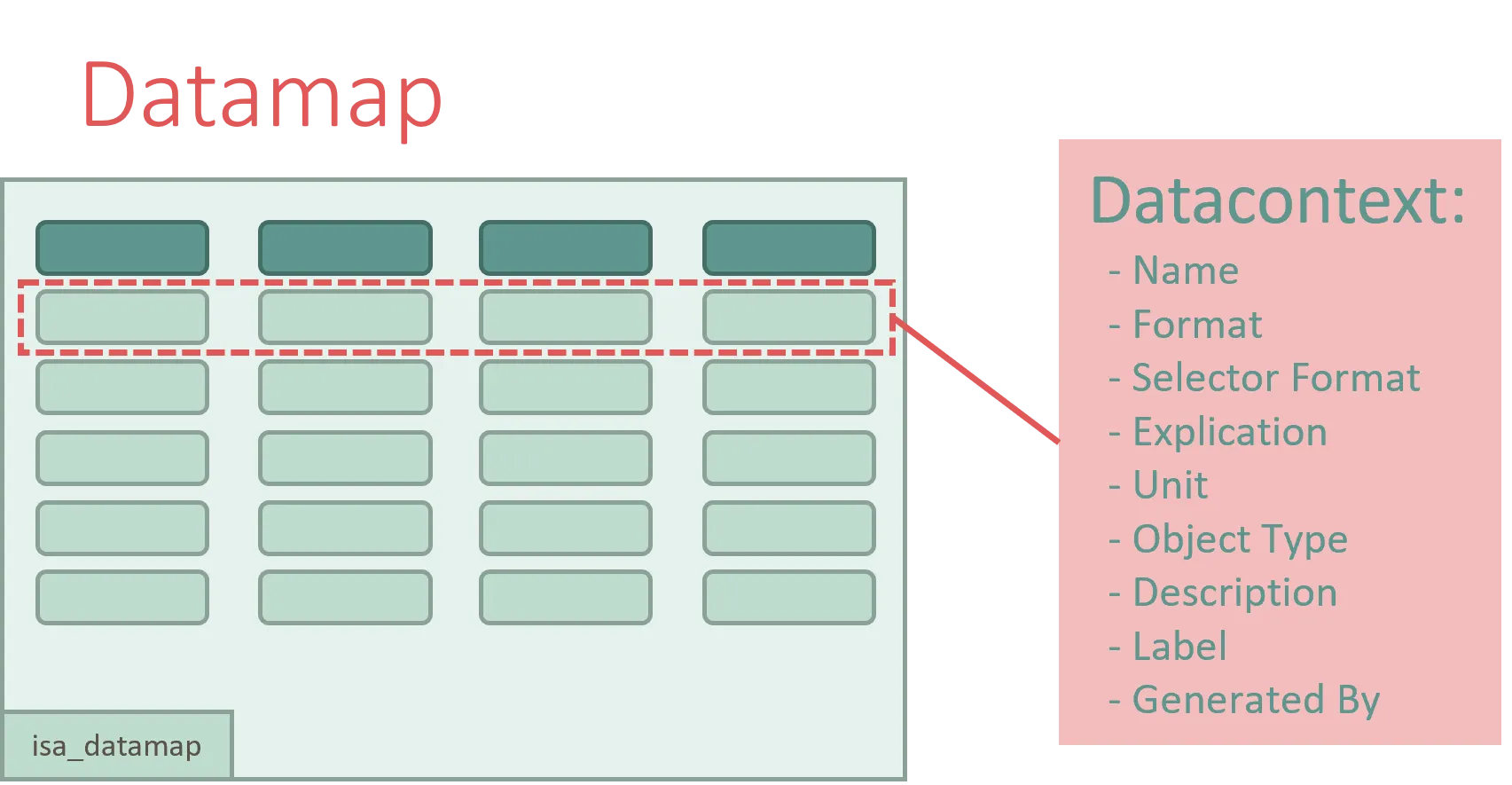
The Datamap itself is a table with fixed columns. Each row in the Datamap represents and annotates a Data Fragment of a data file and is modeled as an DataContext object. The Datamap therefore consists of a collection of DataContext objects.
In this small tutorial we will show how to create a Datamap and integrate it into an ARC. The Datamap will describe a CSV file called sugar_result.csv located in assays/MyAssay/dataset, with two columns, each containing measurements of cellular sugar content:
sugar_result.csv
| sug_s1_r1 | sug_s1_r2 |
|---|---|
| 15.5 | 14.8 |
| 15.2 | 15.1 |
| 15.9 | 14.7 |
First, we will prepare some ontology terms that we will use in the Datamap.
open ARCtrl
let molecule_count = OntologyAnnotation( name="molecule count", tsr ="UO", tan="http://purl.obolibrary.org/obo/UO_0000192")
let mmol_p_kg = OntologyAnnotation( name="Millimole per Kilogram", tsr="NCIT", tan="http://purl.obolibrary.org/obo/NCIT_C68892")
let float_type = OntologyAnnotation( name="float", tsr="NCIT", tan="http://purl.obolibrary.org/obo/NCIT_C48150")// Will be part of tutorial for ARCtrl >=3.0.0from arctrl.arctrl import OntologyAnnotation
molecule_count = OntologyAnnotation( name="molecule count", tsr ="UO", tan="http://purl.obolibrary.org/obo/UO_0000192")
mmol_p_kg = OntologyAnnotation( name="Millimole per Kilogram", tsr="NCIT", tan="http://purl.obolibrary.org/obo/NCIT_C68892")
float_type = OntologyAnnotation( name="float", tsr="NCIT", tan="http://purl.obolibrary.org/obo/NCIT_C48150")We create a Datamap that describes two columns of a CSV file.
Here we create two DataContext objects, which represent each a row in the Datamap and annotate one column in the CSV file.
Afterwards, we will initialize a Datamap object with the two DataContext objects.
let col1Entry = DataContext( name="assays/MyAssay/dataset/sugar_result.csv#col=1", format="text/csv", selectorFormat="https://datatracker.ietf.org/doc/html/rfc7111", explication=molecule_count, unit=mmol_p_kg, objectType=float_type, label="sug_s1_r1", description="Measured sugar concentration in sample 1, replicate 1")
let col2Entry = DataContext( name="assays/MyAssay/dataset/sugar_result.csv#col=2", format="text/csv", selectorFormat="https://datatracker.ietf.org/doc/html/rfc7111", explication=molecule_count, unit=mmol_p_kg, objectType=float_type, label="sug_s1_r2", description="Measured sugar concentration in sample 1, replicate 2")
let datamap = DataMap(ResizeArray[col1Entry; col2Entry])// Will be part of tutorial for ARCtrl >=3.0.0from arctrl.Core.data_map import DataContext, DataMap
col1Entry = DataContext( name="assays/MyAssay/dataset/sugar_result.csv#col=1", format="text/csv", selector_format="https://datatracker.ietf.org/doc/html/rfc7111", explication=molecule_count, unit=mmol_p_kg, object_type=float_type, label="sug_s1_r1", description="Measured sugar concentration in sample 1, replicate 1")
col2Entry = DataContext( name="assays/MyAssay/dataset/sugar_result.csv#col=2", format="text/csv", selector_format="https://datatracker.ietf.org/doc/html/rfc7111", explication=molecule_count, unit=mmol_p_kg, object_type=float_type, label="sug_s1_r2", description="Measured sugar concentration in sample 1, replicate 2")
datamap = DataMap([col1Entry, col2Entry])We have two approaches to write the Datamap to a file:
1. Single File
Using the XlsxController to write the Datamap to a singular Excel file, usable in or out of the ARC context.
open ARCtrl.Spreadsheet
XlsxController.Datamap.toXlsxFile("isa.datamap.xlsx", datamap)// Will be part of tutorial for ARCtrl >=3.0.0from arctrl.arctrl import XlsxController
XlsxController.Datamap().to_xlsx_file("isa.datamap.xlsx", datamap)2. As part of an ARC
Include the Datamap in an ARC and write or update the whole ARC, which will include the Datamap as well.
ARC from a repository.DataMap of the assay “MyAssay” to our datamap.ARC to write the changes back to the repository.let arc = ARC.load("path_to_my_arc")
arc.ISA.Value.GetAssay("MyAssay").DataMap <- Some datamap
arc.Update("path_to_my_arc")// Will be part of tutorial for ARCtrl >=3.0.0from arctrl.arctrl import ARC
arc = ARC.load("path_to_my_arc")
arc.ISA.GetAssay("MyAssay").DataMap = datamap
arc.Update("path_to_my_arc")